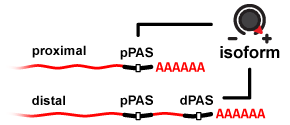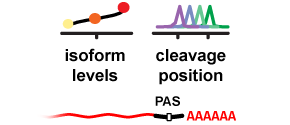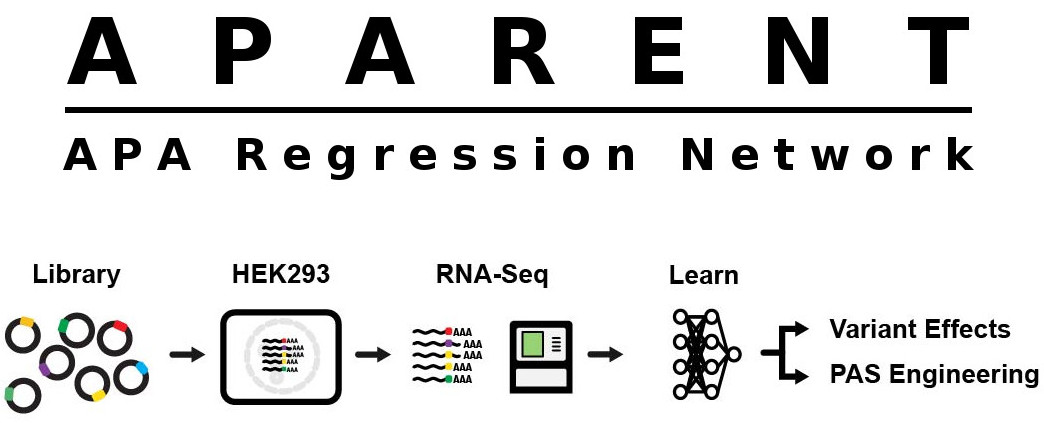
APARENT is a deep neural network that can predict human 3' UTR Alternative Polyadenylation (APA) and the effects of genetic variants on APA. The network was trained on >3.5 million randomized 3' UTR poly-A signals expressed on mini gene reporters in HEK293.
APARENT was described in Bogard et al, Cell 2019 in press.
It was developed in Seelig Lab at the University of Washington.
Contact jlinder2 (at) cs.washington.edu for any questions about the model or data.